JCAST - Junction Centric Alternative Splicing Translator
- Edward Lau Lab, University of Colorado School of Medicine
- Maggie Lam Lab, University of Colorado School of Medicine

About JCAST
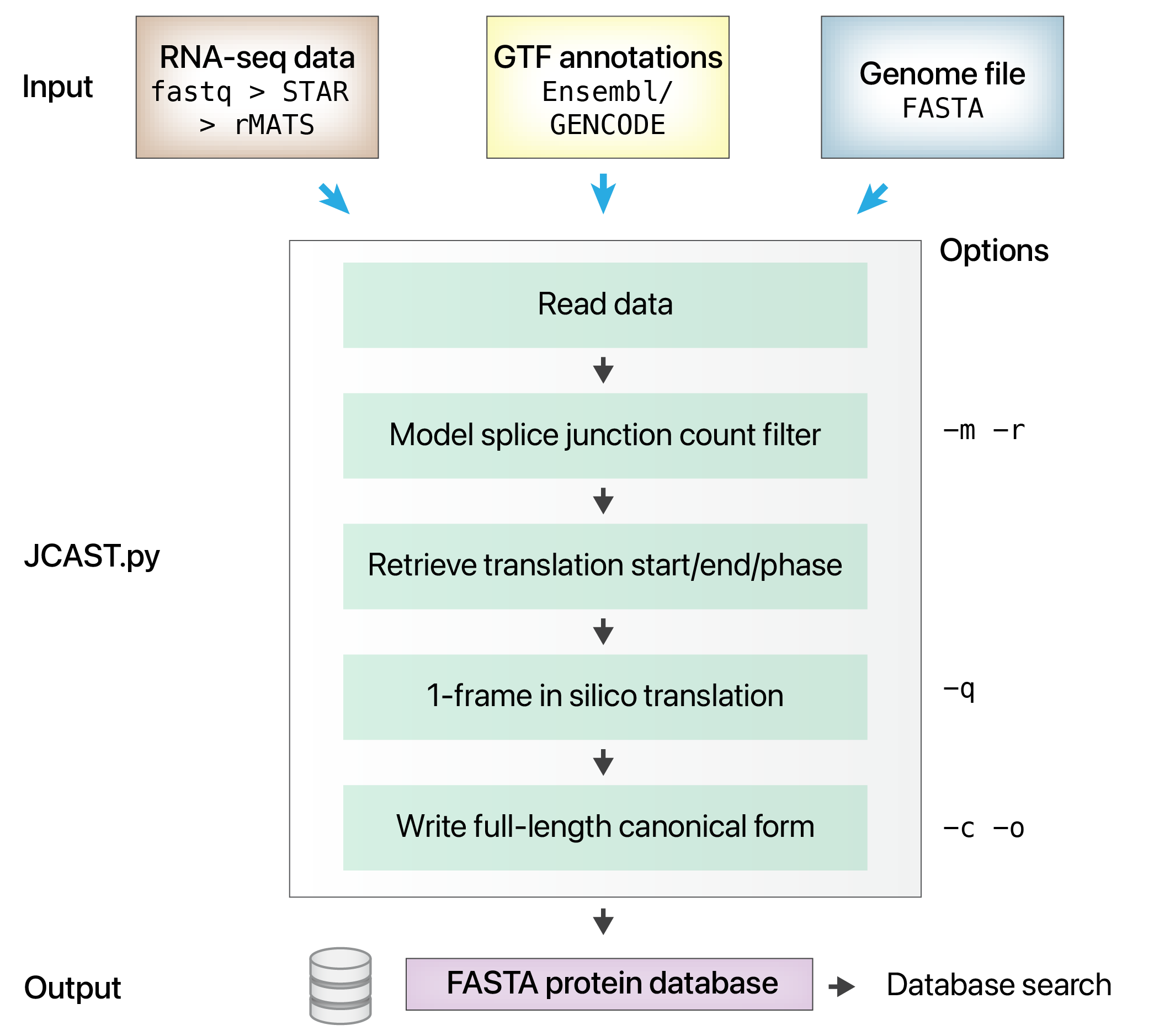
JCAST (Junction Centric Alternative Splicing Translator) is a Python software that takes in splicing events from RNA sequencing data to create custom protein sequence databases for protein isoform analysis.
JCAST attempts to create small protein sequence databases that contain only likely sequences in mass spectrometry experiment samples to avoid runaway database size inflation.
To do so, JCAST likely translatable alternative splice junctions and performs one-frame in silico analysis, then joins the translated variant sequences with full-length canonical sequences.
Downloads
Latest Updates
v0.3.5a
- JCAST will now skip a junction if the nucleotide sequence contains masked bases
- Added the –mask argument to determine masked nucleotide letters (default: NnXx)
- Added an option to translate only a subset of splice types (MXE, RI, SE, A3SS, A5SS) with the -s argument
v0.3.5
- Fixed an issue where a small number of slices were not translated.
- Fixed an issue where JCAST quits if the model flag is not set.
See Change Log for details.
The latest version and source code of JCAST can be found on github: https://github.com/ed-lau/jcast.
See the Quick Start and Documentation for instructions.
Contributors
- Edward Lau, PhD - ed-lau
- Maggie Lam, PhD - Maggie-Lam
- R W Ludwig, BSc - WesLudwig
Citations
JCAST: Sample-Specific Protein Isoform Databases for Mass Spectrometry-based Proteomics Experiments. RW Ludwig, E Lau Software Impacts 10, 100163 Link
Determining Alternative Protein Isoform Expression Using RNA Sequencing and Mass Spectrometry. Y Han, JM Wright, E Lau, MPY Lam STAR Protocols 1 (3), 100138 Link
Splice-Junction-based Mapping of Alternative Isoforms in the Human Proteome. E Lau, Y Han, DR Williams, CT Thomas, R Shrestha, JC Wu, MPY Lam Cell Reports 29 (11), 3751-3765. e5 Link